Single cell analysis with scanpy
Installation
I recommend using Miniconda to manage packages and creating an entirely new environment to install scanpy.
conda install -c conda-forge scanpy python-igraph leidenalgBasic conception before your start
AnnData class
An "AnnData" class is the interface to the Scanpy operation, meaning we must create an AnnData object as a parameter to the Scanpy function.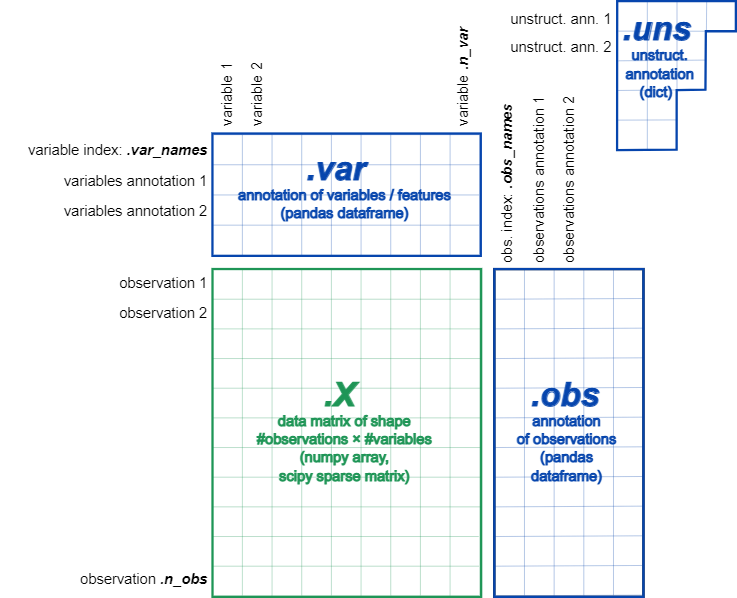
At the most basic level, an AnnData object adata stores a data matrix adata.X, annotation of observations adata.obs and variables adata.var as pd.DataFrame and unstructured annotation adata.uns as dict. Names of observations and variables can be accessed via adata.obs_names and adata.var_names, respectively. AnnData objects can be sliced like dataframes, for example, adata_subset = adata[:, list_of_gene_names]
import scanpy as sc
import os
adata = sc.read_csv('../data/GSM5226574_C51ctr_raw_counts.csv').T
adataLet's see the properties of AnnData.
type(adata)
type(adata.X)
adata.var #extract var(python DataFrame object that describe genes)
adata.obs #extract observations(python DataFrame object that describe cells)
adata.var_names #return an index objects of gene names
adata.obs_names #return an index objects of cell barcodes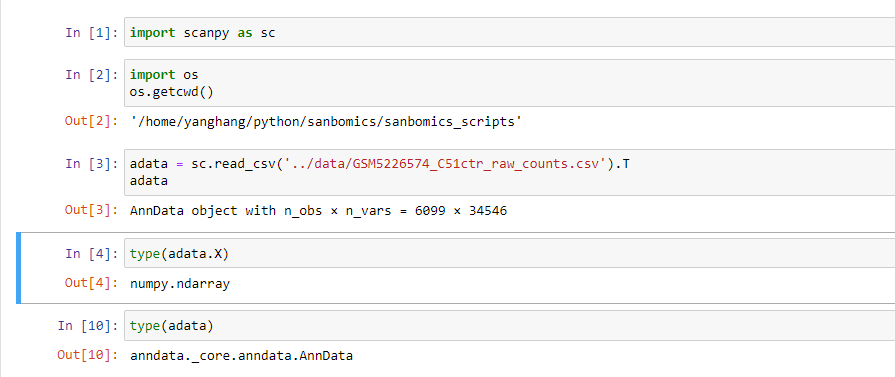
案例丰富且贴合主题,论证逻辑环环相扣。
跨文化对比分析视角值得深入探索。
作者的观点新颖且实用,让人在阅读中获得了新的思考和灵感。